10.4: Diésteres de fosfato
- Page ID
- 2387
10.4A: Phosphate diesters as the backbone for DNA and RNA
Phosphate diesters play an absolutely critical role in nature - they are the molecular 'tape' that connect the individual nucleotides in DNA and RNA.
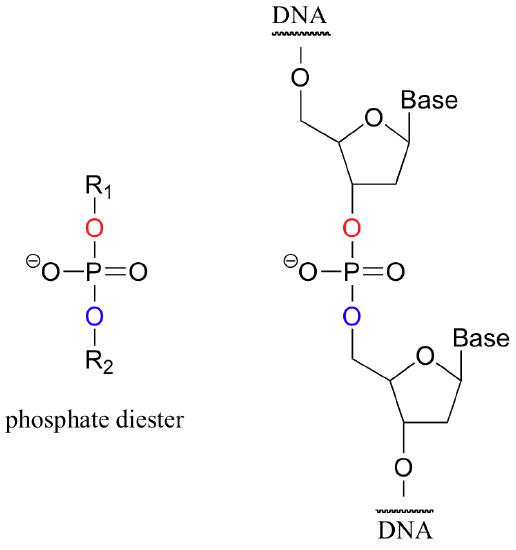
In a very interesting essay that is well worth reading, F.H. Westheimer of Harvard addressed the question of why phosphates were chosen by nature for this critical biochemical job (Science 1987, 235, 1173). He pointed out that other molecules could hypothetically perform the same function: citrate, for example, might have been used to link nucleotides with ester functional groups.
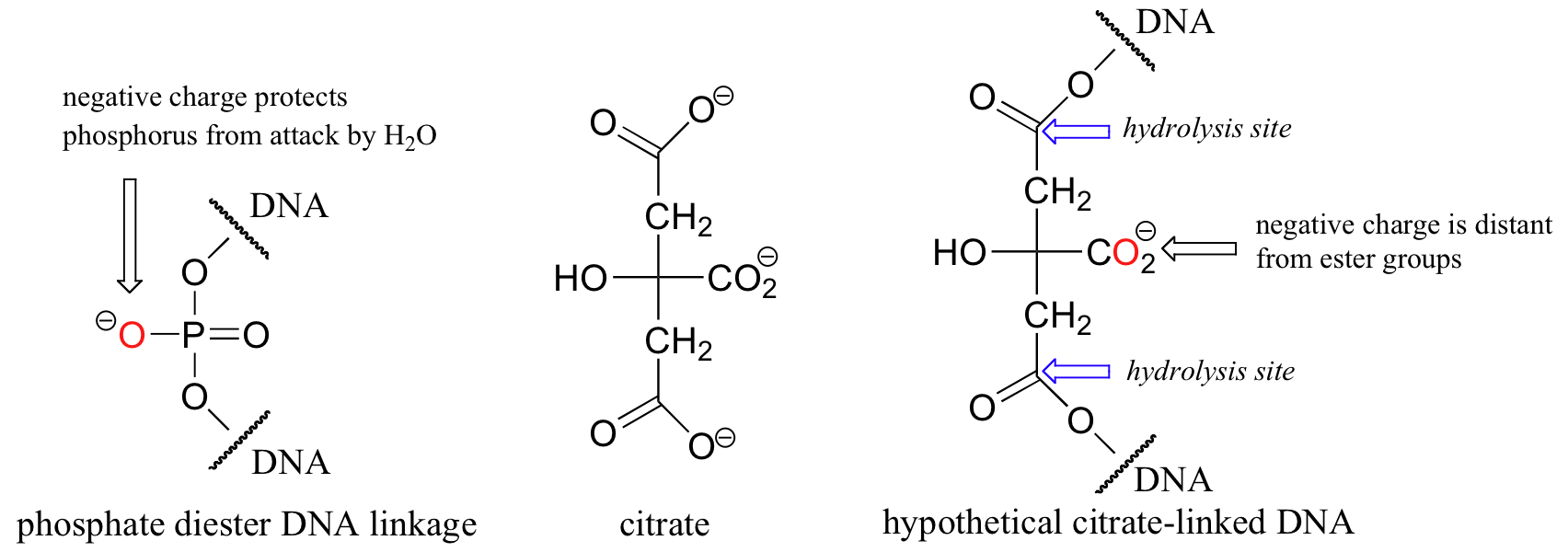
Westheimer argued that such ester-linked DNA would be far too unstable to support life, as the rate of spontaneous (non-enzymatic) hydrolytic breakdown for esters is unacceptably high: all it takes is a single break in the DNA chain to potentially prevent an organism from passing along its genes to the next generation. (The hydrolysis of esters is a reaction that we will examine in detail in section 12.4B). Phosphate diesters, on the other hand, are much more resistant to spontaneous hydrolysis, with rate constants approximately 5 million times slower than carboxylate esters. The main source of this stability is the negative charge on the non-bridging oxygen. This negative charge effectively repels potential nucleophilic water molecules, shielding the phosphorus atom from attack. While the hypothetical citrate diester-linked DNA also has negatively charged group (a carboxylate) available to carry out the same function, this negative charge is much further away from the two possible sites of hydrolysis, and thus is not nearly as effective a shield.
While DNA is inherently resistant to spontaneous hydrolysis, RNA is quite vulnerable to hydrolytic breakdown, even in aqueous solutions carefully buffered to neutral pH. This does not present a physiological dilemma, because the function of RNA is to temporarily transmit genetic information, while DNA serves as permanent information storage for the lifetime of the organism. Why does hydrolysis occur so much more rapidly in RNA than in DNA? The answer is on the 2' carbon: RNA nucleotides, unlike the 2'-deoxynucleotides of DNA, have a hydroxyl group here. This 2' hydroxyl group is poised in a position where it can attack the phosphorus of the phosphate diester, breaking the DNA chain and forming a cyclic phosphate diester, which is subsequently opened by hydrolysis.
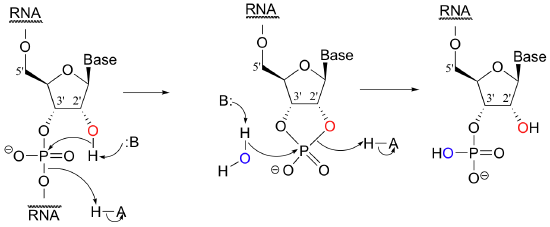
Researchers working with RNA have to be careful to store their samples at very cold temperatures, preferably freeze-dried or precipitated in ethanol, to avoid hydrolysis. The problem of RNA decomposition is compounded by the fact that RNAase enzymes, which catalyze RNA hydrolysis, are present on the surface of human skin and are very stable, long-lived, and difficult to destroy.
10.4B: The organic chemistry of genetic engineering
Many enzymes that catalyze reactions involving the phosphate diester bonds of DNA have been harnessed by genetic engineers - scientists who copy, snip, and splice DNA in order to create custom versions of genes. The tools of genetic engineering have become indispensable and commonplace in the past decade, and most researchers working on the biological side of chemistry use them extensively. The days of painstakingly purifying an enzyme from bacterial cultures or ground up cow livers are pretty much gone. Now scientists clone the gene that encodes the enzyme, make any desired changes (by site-directed mutagenesis, for example), and use a host such as E. coli or yeast to produce the enzyme from the cloned gene. You will learn the details of many of these procedures in a biochemistry or molecular biology course. What we will focus on now are a few of the key organic reactions that are involved.
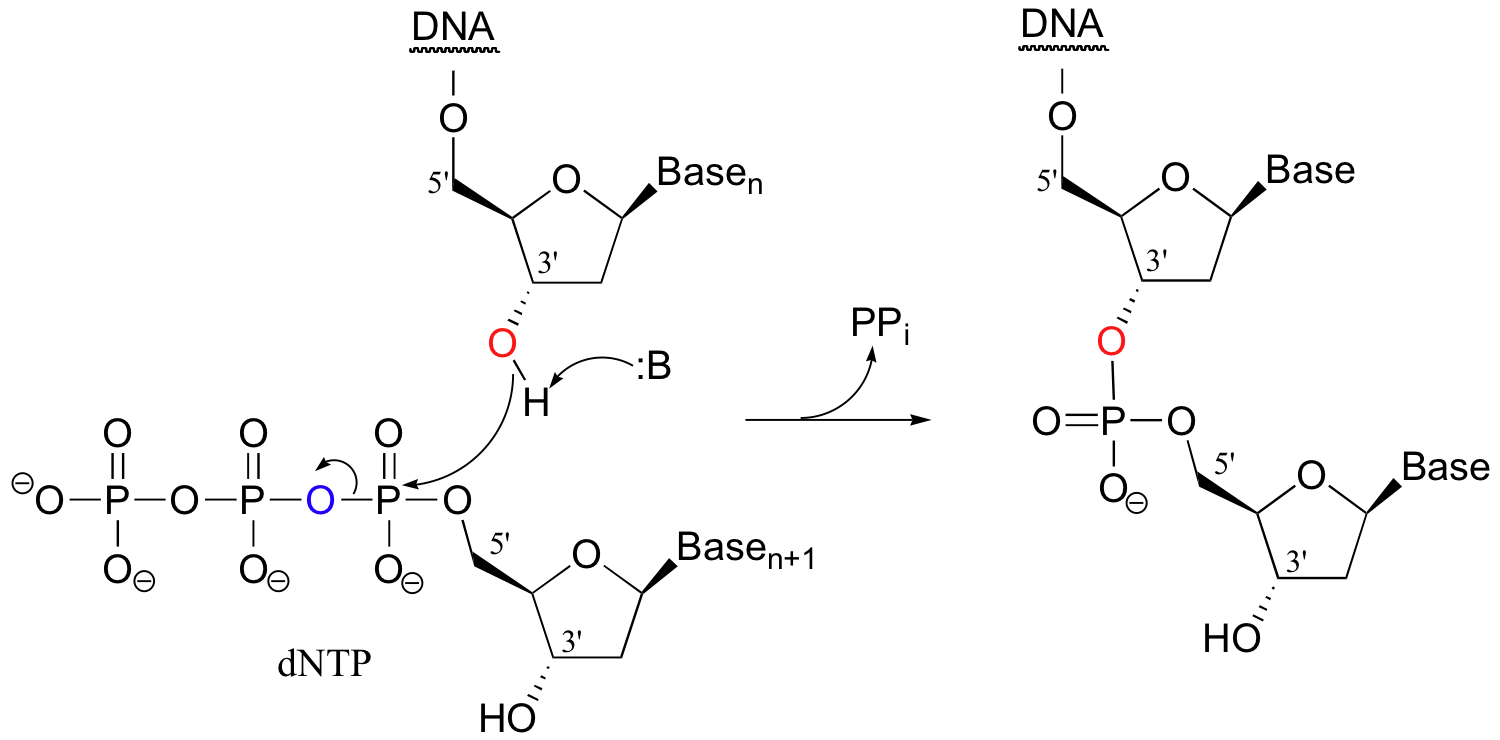
Often the first thing you have to do in a genetic engineering procedure is to copy a DNA strand. This is accomplished by an enzyme called DNA polymerase, which uses a single strand of DNA as a template to synthesize a second, complementary strand (the biochemical details of this complex process are beyond the scope of this text). In the polymerase reaction, the 3' hydroxyl group on the end of the growing DNA strand attacks the a-phosphate of a 2'-deoxynucleoside triphosphate, expelling inorganic pyrophosphate.
Scientists are able to cut DNA using 'molecular scissor' enzymes called restriction endonucleases that cleave double-stranded DNA at specific base sequences. The chemistry is conceptually simple - it is merely the catalyzed hydrolysis of a phosphate diester.
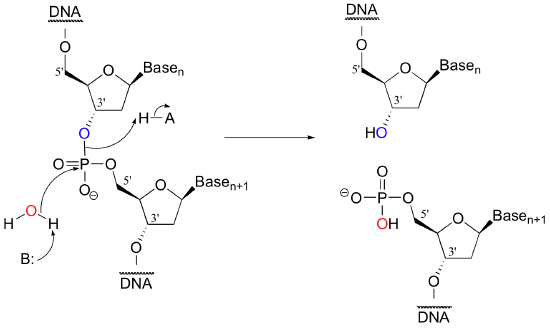
Notice that the result of this hydrolytic cleavage reaction is one segment of DNA with a hydroxy group at the 3' position, and a second segment with a phosphate group at the 5' position.
A commonly used restriction endonuclease called 'BamHI' makes cuts specifically at the following 6-base sequence:
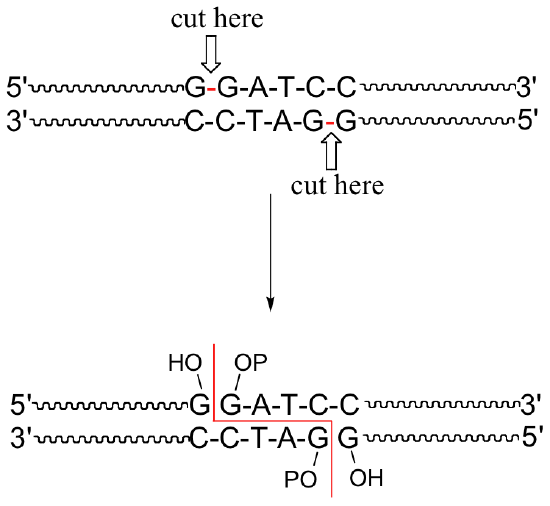
Notice that a 'staggered' cut is made: this is a common (and useful) property of many endonucleases, although some make 'blunt-ended' cuts.
The reverse of an endonuclease-catalyzed phosphate diester hydrolysis is the reaction catalyzed by an enzyme called DNA ligase. In a ligation reaction, the 3' hydroxyl and 5' phosphate ends of two DNA strands are tied together through the formation of a new phosphate diester linkage.
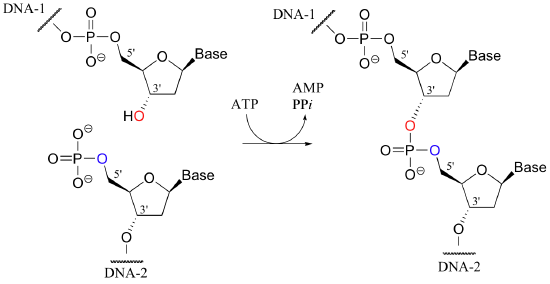
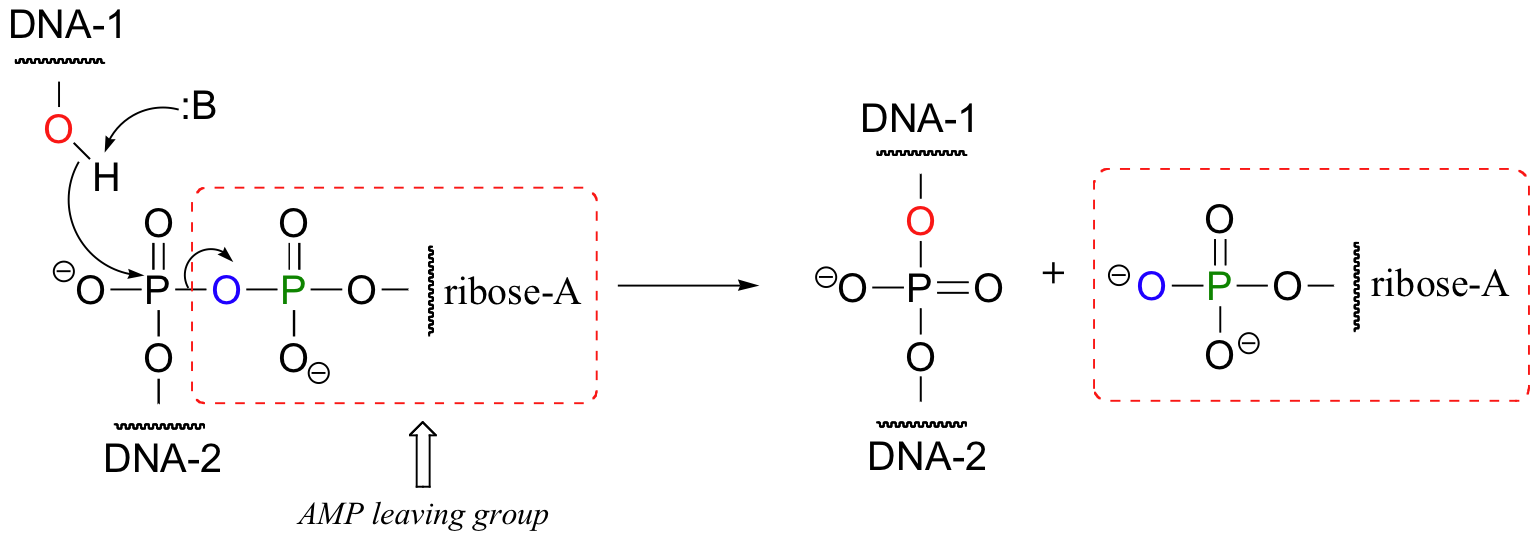
This is a thermodynamically uphill transformation, so energy from ATP must be 'spent' in order to drive it forward. In the first step of the reaction, an AMP group is transferred from ATP to a lysine residue on the ligase enzyme, forming a phosphoamide bond (this is the first example we have seen in which a phosphoryl group is transferred to anything other than an oxygen).

The enzyme next transfers the AMP group to the phosphorylated 5' end of a DNA strand, forming a new phosphate anhydride bond:
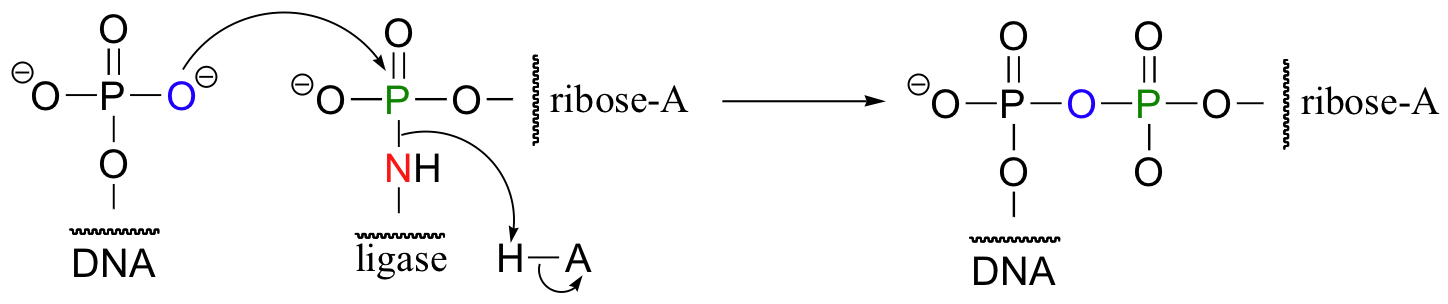
What has happened here is that the negatively-charged oxygen of the phosphate, a very poor leaving group, has been transformed into an (excellent) AMP leaving group, at the expense of one ATP. The hydroxyl group on the 3' end of the DNA strand can now attack the 5' phosphate, driving off the AMP:
One more enzymatic tool in the genetic engineering arsenal bears mention. In some situations, a researcher may want to prevent ligation from occurring in a particular DNA sample. This can be accomplished by using the enzyme alkaline phosphatase, which catalyzes the dephosphorylation of many organic monophosphate esters, including 5'-phosphorylated DNA (recall that we discussed phosphatases in section 10.3)
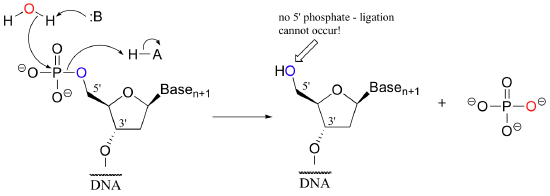
With the phosphate group removed, ligation is impossible - there is no way to make a new phosphodiester bond without a phosphate group! Alkaline phosphatase only hydrolyzes monoesters, not diesters, so the DNA strand is otherwise untouched.


