12.6: Reacciones de sustitución nucleofílica de acilo que implican enlaces peptídicos
- Page ID
- 2405
12.5A: Formation of peptide bonds on the ribosome
You are probably quite familiar, from your biology classes and from chapter 6 of this text, with the basic structure of the peptide bonds that connect the amino acids in proteins. Peptide bonds are simply amide functional groups that link the amino acids in a protein.
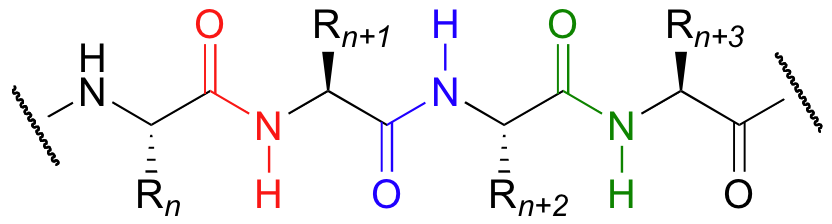
Let’s take a look at the chemistry behind the formation of new peptide bonds in a growing protein molecule. This process takes place on the ribosome, which is basically a large biochemical 'factory' for the construction of proteins. You will learn many more details in other classes about the complex but fascinating process of ribosomal protein synthesis. For now, we will concentrate on the relatively simple organic transformation that is taking place: the formation of an amide starting from a carboxylate and an amine.
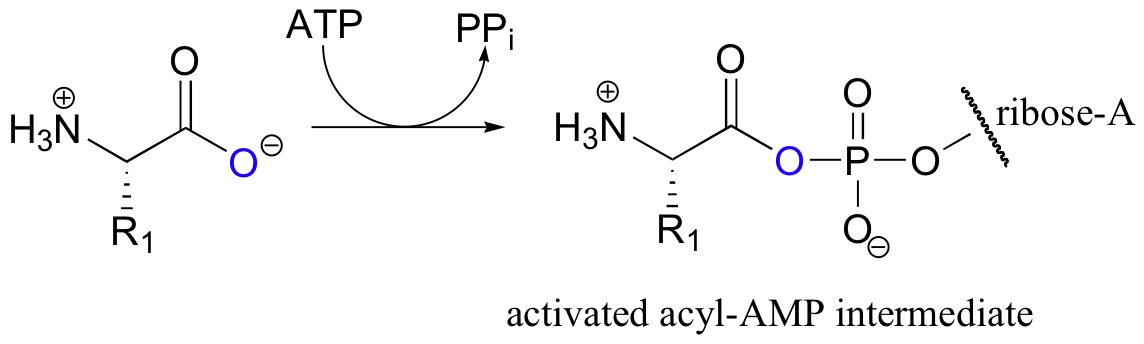
We have seen this same functional group transformation before – think back to the glutamine and asparagine synthetase reactions (section 12.2). The same ideas that we learned for those reactions hold true for peptide bond formation: the carboxylate group on a substrate amino acid must first be activated, and the energy for this activation comes from ATP. As with asparagine synthetase, the carboxylate group of the amino acid is first transformed to an acyl-AMP intermediate through a nucleophilic substitution reaction at the alpha-phosphate of ATP.
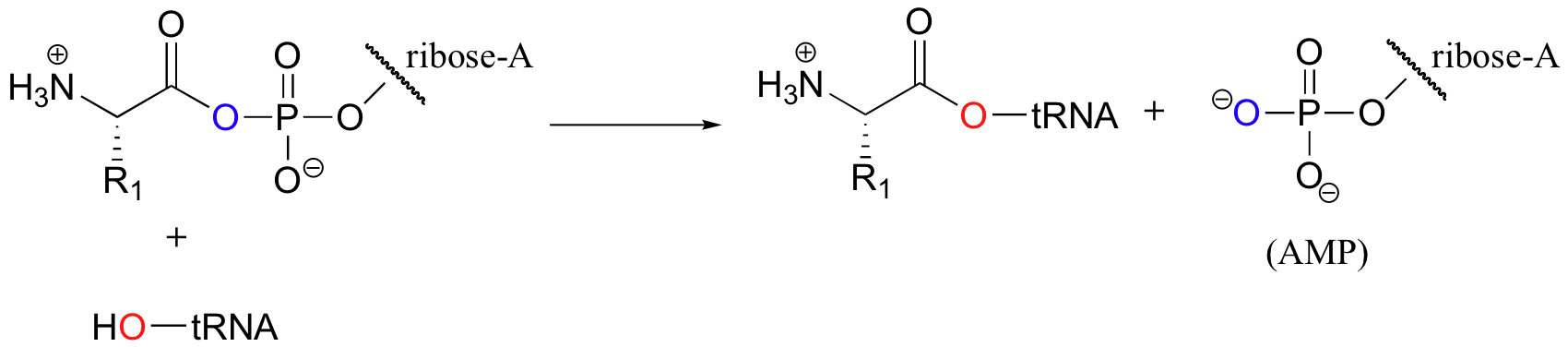
In the next step, the amino acid is transferred to a special kind of RNA polymer called transfer RNA, or tRNA for short. We need not concern ourselves here with the structure of tRNA molecules- all we need to know for now is that the nucleophile in this reaction is a hydroxyl group on the terminal adenosine of tRNA.

The incoming nucleophile is an alcohol, thus what we are seeing is an esterification: an acyl substitution reaction between an activated carboxylate (specifically an acyl-AMP) and an alcohol to form an ester.
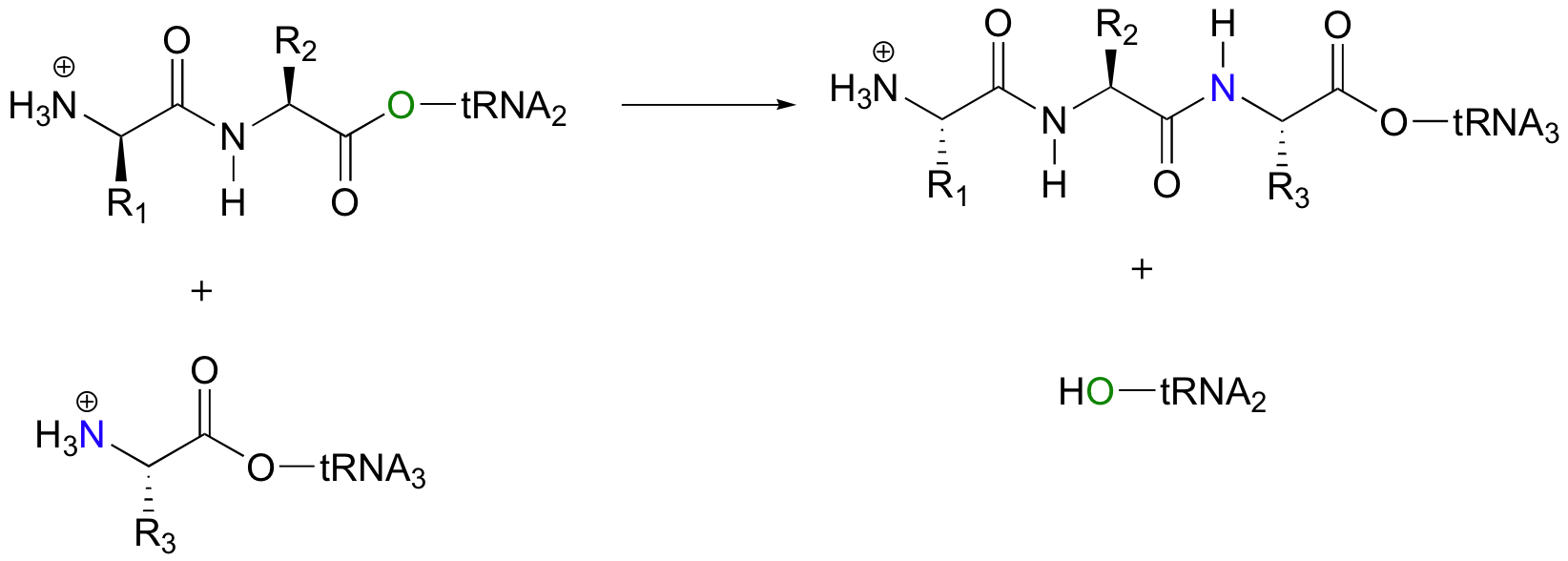
The first amino acid is now linked via an ester group to the tRNA molecule. The actual peptide bond-forming reaction occurs when a second amino acid, also linked to its own tRNA molecule, is positioned next to the first amino acid. In another acyl substitution reaction, the amino group on the second amino acid displaces the tRNA on the first amino acid – an ester has been converted to an amide (thermodynamically downhill, so ATP is not required).

This process continues on the ribosome, as one amino acid after another is added to the growing protein chain:
When a genetically-coded signal indicates that the chain is complete, an ester hydrolysis reaction – as opposed to another amide formation - occurs on the last amino acid.

This frees the mature protein from the ribosome, and results in the formation of a free carboxylate group at the end of the protein (this is called the carboxy-terminus, or C-terminus of the protein, while the other end – the ‘starting’ end – is called the N-terminus).
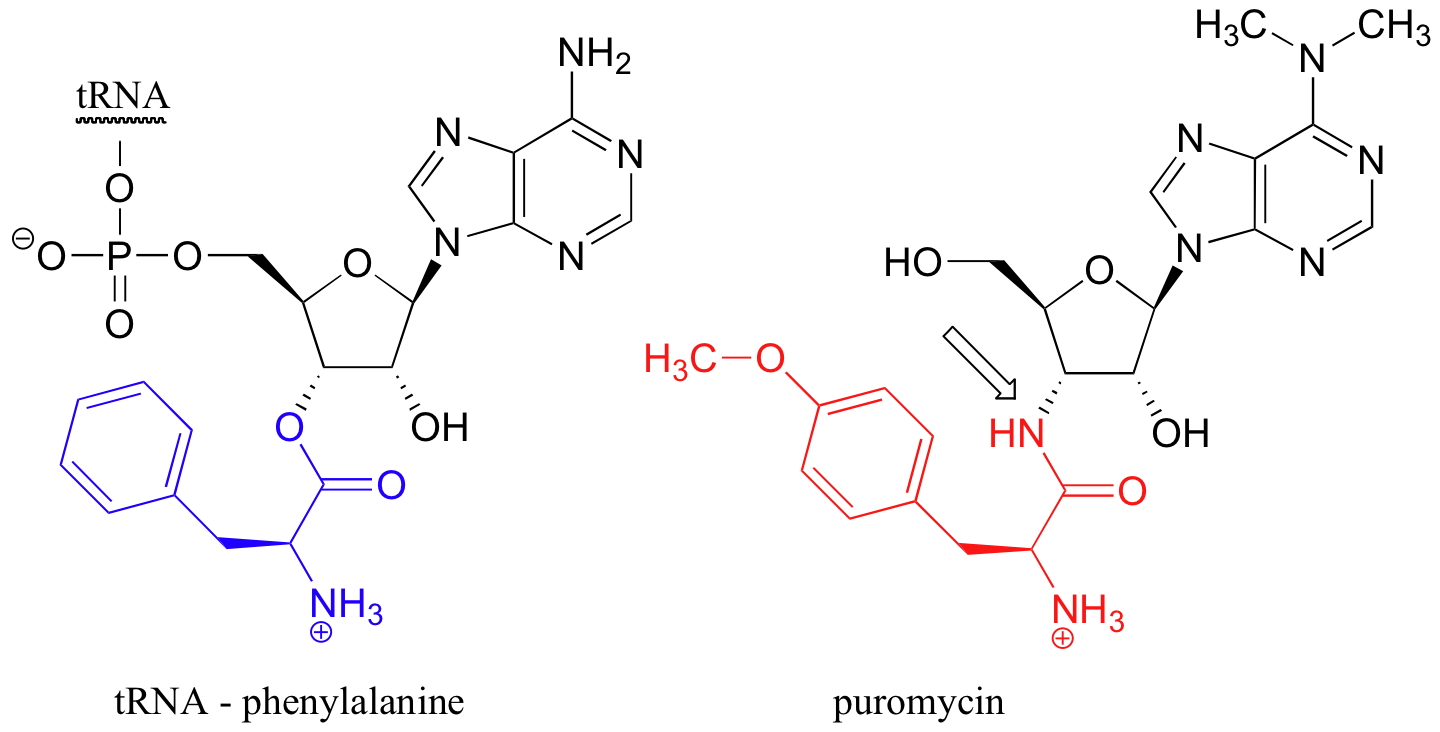
Many potent drugs work by interfering with the protein synthesis process. The antibiotic puromycin, for example, mimics the structure of a phenylalanine amino acid attached to the terminal adenosine of tRNA, with an amide linkage (see arrow) where there would normally by an ester. Puromycin causes premature termination of protein synthesis when the phenylalanine mimic part of the molecule (in red below) adds to a growing peptide chain, but then is not released from the tRNA-mimic part (in black) due to the stability of the amide (as opposed to ester) linkage.
12.5B: Hydrolysis of peptide bonds - HIV protease
Peptide bonds in proteins and polypeptides are subject to spontaneous (nonenzymatic) hydrolysis in water, but although this amide to carboxylate conversion is thermodynamically a downhill reaction, it is kinetically very slow at neutral pH (look back at Chapter 6 if you need to review the difference between the thermodynamics and kinetics of a reaction). In fact, the half-life for uncatalyzed hydrolysis of a peptide bond in pH 7 water is estimated to be as high as 1000 years. The kinetic stability of peptide bonds towards uncatalyzed hydrolysis can best be explained by a resonance structure that contains a carbon-nitrogen double bond and a separation of charge.
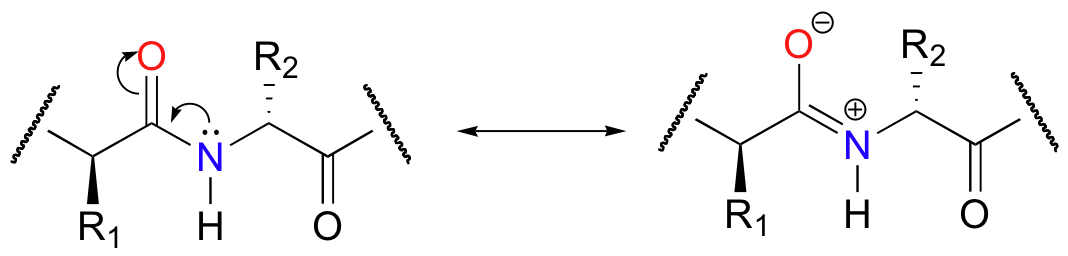
This partial double bond character implies that the carbon-nitrogen bond is relatively strong, and also that the carbonyl carbon is less electrophilic towards attack by a water molecule.
The stability of peptides bonds makes good physiological sense: we would all be in trouble if our enzymes, receptors, and structural proteins were hydrolyzing away while we slept! That being said, it is also true that controlled, specific hydrolysis of peptide bonds, catalyzed by a large, diverse class of enzymes called proteases, is a critical biochemical reaction type that often occurs very rapidly. Proteins that we eat must be hydrolyzed so that small polypeptides and individual amino acids can be absorbed. Also, many enzymes are only active after they have been ‘cut’ (hydrolyzed) at a specific amino acid location by a protease.
Although all proteases catalyze essentially the same reaction – amide hydrolysis - different protease subfamilies have evolved different catalytic strategies to accomplish the same result. Among the most well-studied enzymatic reactions in biochemistry is that catalyzed by chymotrypsin, a member of the 'serine protease' subfamily. This enzyme specifically hydrolyzes peptide bonds on the C-terminal side of large, hydrophobic amino acids such as phenylalanine, tyrosine, and tryptophan.
Because the mechanism of the chymotrypsin reaction is explained in great detail in most biochemistry textbooks, we will not discuss it in depth here. Instead, let's look at an example of a different protease subfamily. HIV protease is the target of some the most recently-developed anti-HIV drugs. It plays a critical role in the life cycle if the HIV virus, hydrolyzing specific peptide bonds of essential viral proteins in order to convert them to their active forms. HIV protease is a member of the aspartyl protease subfamily, so-named because of the two aspartate residues located in the active sites of these enzymes.
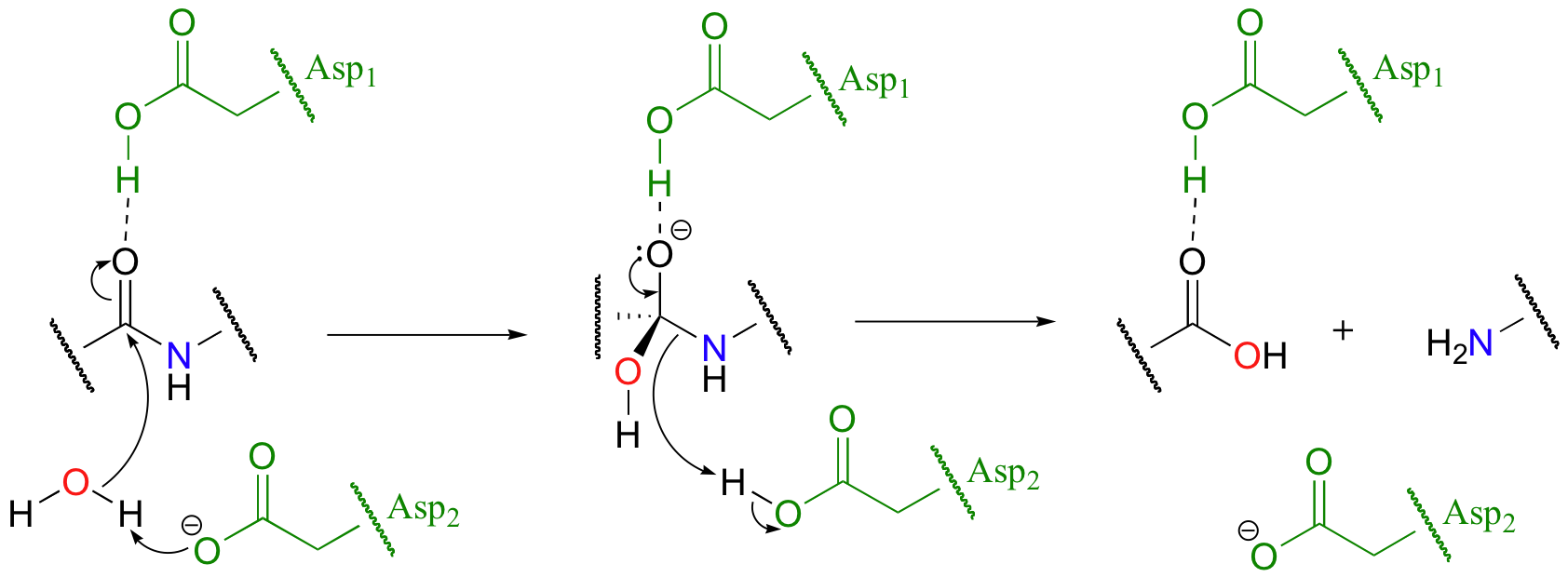
In aspartyl proteases, the two enzymatic aspartates work in concert to activate the electrophile, nucleophile, and leaving group in the reaction. Exactly how this works is a subject of some debate and the details may well vary according to the enzyme in question, but one likely mechanism is illustrated in the figure above. Recently, a crystal structure of an HIV protease lead to a proposal for a slight variation on this theme - see Biochemistry 2007, 46, 14854, fig 8 for details.
12.5C: The chemical mechanism of penicillin
The discovery of penicillin was arguably one of the most important events in the history of modern medicine. Penicillin, and later generations of antibiotic drugs, have saved countless lives from once-deadly bacterial infections. The elucidation of the chemical mechanism of penicillin action was also a milestone in our developing understanding of how drugs function on a molecular level. We now know that penicillin, and closely related drugs such as ampicillin and amoxycillin (see section 1.4C), work by irreversibly inhibiting an enzyme that is involved in the construction of bacterial cell walls. These cell walls are composed of 'peptidoglycan': sugar chains crosslinked by short polypeptides. The enzyme targeted by penicillin is a transpeptidase, which connects two short polypeptides through the formation of a new peptide bond between an alanine and a glycine. Like many other reactions involving peptide bonds, the first step of the transpeptidase reaction is the formation of an ester link between one of the polypeptide substrates and a serine on the enzyme. In this case, the terminal alanine of the polypeptide substrate is expelled.
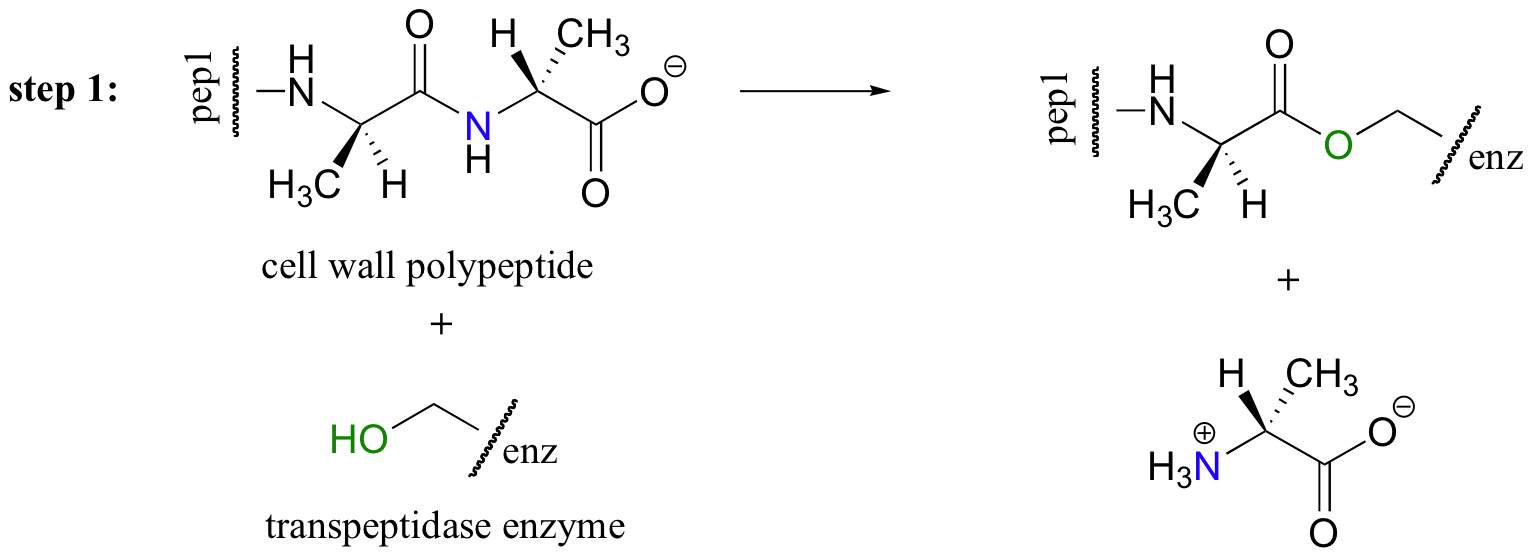
In the second step, the N-terminal glycine of the second short polypeptide substrate displaces the enzymatic serine to form a new peptide (amide) bond:

Penicillin, which in its lowest-energy conformation mimics the natural polypeptide substrate well enough to be allowed into the enzyme’s active site, serves as an irreversible inhibitor, an alternate target for the nucleophilic serine in the transpeptidase active site.
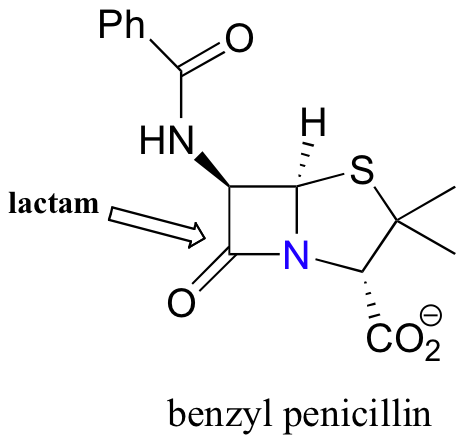
More specifically, it is the cyclic amide (also called a lactam) group that is the target. Because of the high degree of ring strain in the four-membered ring, a large amount of energy is released when the lactam is broken apart and the ring opened up – in other words, the ring is a compressed spring just waiting to be released by means of an acyl substitution reaction. When the nucleophilic serine attacks and opens the ring, the result is a new covalent linkage between the enzyme and the penicillin molecule, which irreversibly inhibits the normal function of the enzyme and kills the bacterial cell.
Bacteria which are resistant to penicillin and related antibiotics have acquired an enzyme, called beta-lactamase, that catalyzes the hydrolysis of the lactam ring, rendering the drug inactive.
Exercise 12.9: Draw the structure of enzyme-penicillin adduct described above.


