9.6: Problems for Chapter 9
- Page ID
- 2382
Link to Solution Manual
P9.1: Draw a complete curved arrow mechanism for the following reaction. Assume an SN1 mechanism, and that the leaving group leaves as an anion, then is protonated in a later step. Show how both the leaving group and the carbocation intermediate are stabilized by resonance.)
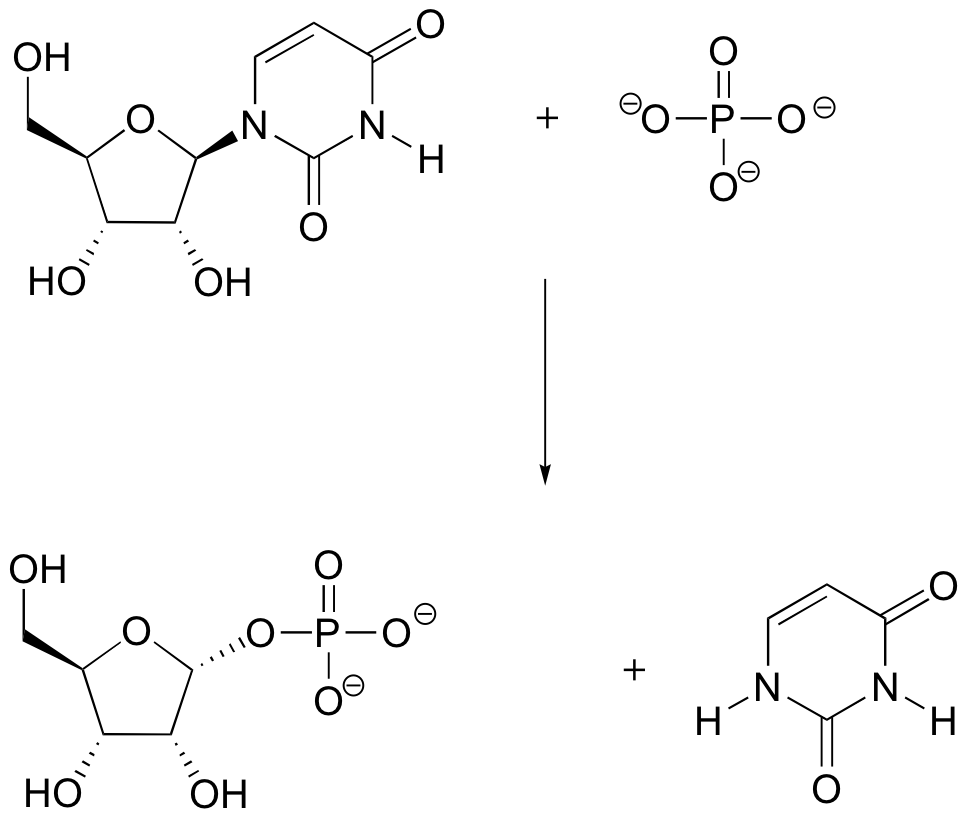
P9.2: The guanine base of DNA can be oxidized to 8-oxoguanine by the so-called 'reactive oxygen species' (ROS) that are constantly being produced in our bodies as a byproduct of respiration (ROS are discussed briefly in section 17.2, as well as in most biochemistry textbooks).
One way that our cells deal with this damaged DNA is to first remove the 8-oxoG base from the sugar-phosphate DNA chain. This is thought to occur with the help of a nucleophilic lysine residue in a type of DNA repair enzyme called DNA glycosylase, a reaction that results in a temporary enzyme-DNA covalent bond. Propose a likely a mechanism for this reaction.
P9.3: Choline, an important neutotransmitter in the nervous system, is formed directly from 2-(N,N-Dimethylamino)ethanol:

a) Besides the enzyme and the starting compound, what other important biomolecule do you expect to play a part in the reaction?
b) Show a mechanism for the reaction.
P9.4: In many extracellular proteins (proteins that are located outside cells) , the sulfur atoms of two cysteine side chains become covalently linked to form what are called ‘disulfide bonds’ (these bonds will be discussed in more detail in section 16.12)
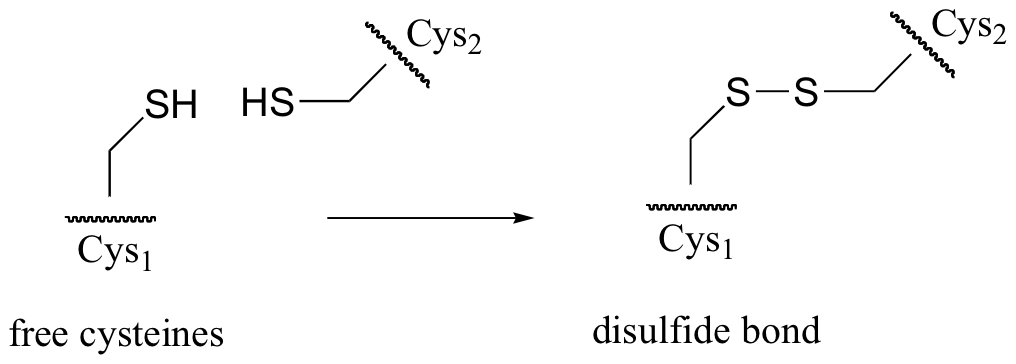
Disulfide linkages are converted back to free cysteines in living cells by the action of a molecule called glutathione.
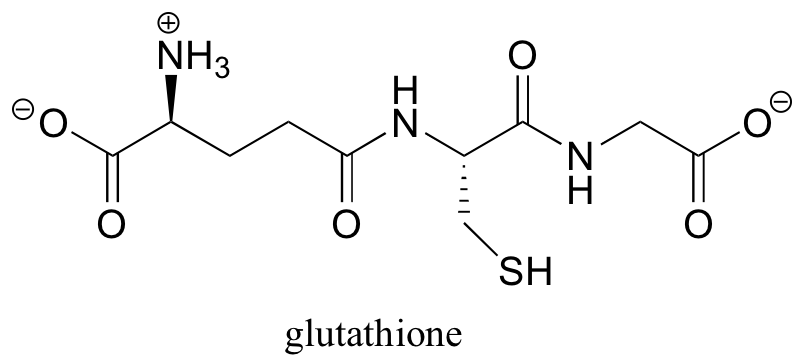
In a spontaneous (uncatalyzed) reaction, the cysteine-cysteine disulfide bond in the protein is broken, and a new disulfide bond forms between two glutathione molecules. This process involves two successive SN2-like reactions (although the electrophiles are not carbons).
a) What do you predict is the key functional group in the action of glutathione?
b) Using appropriate abbreviations, propose a mechanism for this process.
P9.5: The final step in the biosynthesis of the amino acid methionine is dependent on two coenzymes: cobalamin, which is derived from vitamin B12, and 5-methyltetrahydrofolate, a derivative of folic acid. Cobalamine is a large and complex molecule, containing a cobalt metal center that is bound to 5 nitrogen-containing groups.
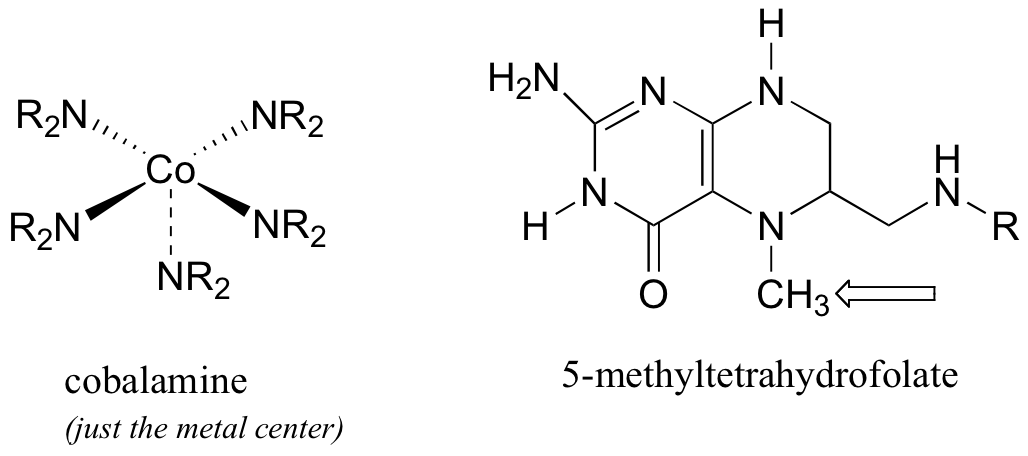
The cobalt metal in cobalamin is an excellent nucleophile. In the methionine biosynthesis reaction, the 5-methyl group from 5-methyltetrahydrofolate ends up on methionine, with overall retention of stereochemical configuration about the methyl carbon (this can be demonstrated by using 2H- and 3H-labels on the methyl group- see section 9.1A).. Propose a likely mechanism for this process, taking into account the stereochemical outcome (you will need to infer the precursor to methionine).
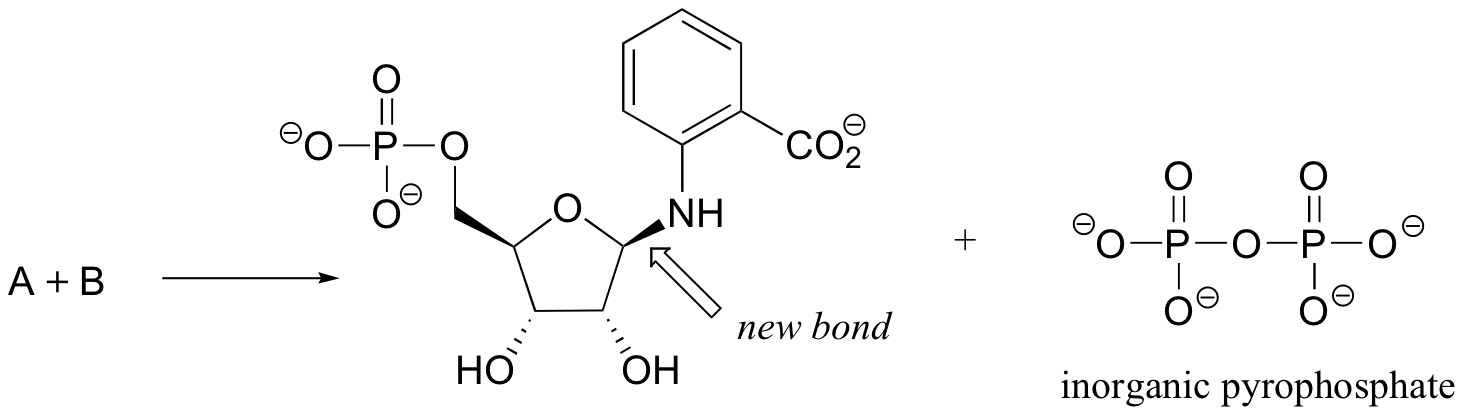
P9.6: The biosynthesis of tryptophan involves the formation of the compound whose structure is shown below. The new bond formed in the reaction is indicated by the arrow, and the reaction proceeds with retention of configuration at the anomeric carbon. Inorganic pyrophosphate is released. Show the structures of the two starting compounds A and B.
Challenge problems
C9.1: An enzyme found in bacteria catalyzes the hydrolysis of mannose sugar-phosphate (the phosphate group is actually GDP).
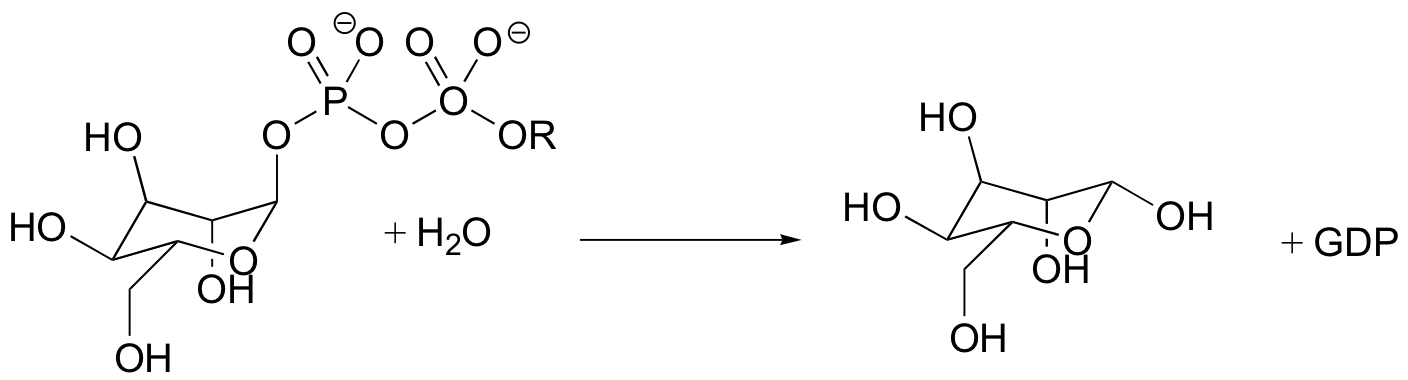
X-ray crystal structure of the enzyme's active site shows an aspartate residue located close to the anomeric carbon of the bound substrate. Site-directed mutagenesis of this aspartate to asparagine leads to a (10 2.6)-fold decrease in the maximal rate of the reaction. In addition, the crystal structure shows that the diphosphate of GDP interacts with three active site arginines, a tyrosine, and a bound magnesium ion (Mg+2) is required for activity.
a) Does this information, taken together, lead you to predict the mechanism to be more dissociative or more associative in character? Explain your prediction.
b) Suggest a fluorine substituted substrate analog that could be used to further address the associative/dissociative character of the reaction.
c) How might using D2O in the reaction buffer further clarify the situation?
C9.2: Glutathione S-transferases (GSTs) are a family of enzymes that use the multifunctional coenzyme glutathione to break down potentially harmful foreign (often referred to as 'xenobiotic' ) compounds in the cell through nucleophilic attack. Predict the likely product(s) of the GST-catalyzed reaction between glutathione and the aromatic epoxide shown below, and show the relevant mechanism.
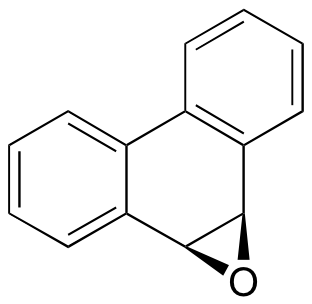
C9.3: A glutathione S-transferase enzyme is thought to be involved in conferring resistance in some bacteria to the antibiotic fosfomycin.
a) Draw a mechanism showing how this GST enzyme could inactivate fosfomycin.
b) The enzyme requires Mg+2 for activity. Propose two possible catalytic roles for the metal ion.
C9.4: Many bacteria have 'dehalogenase' enzymes, which are able to remove halogens from halogenated organic pollutants. This property makes them potentially very useful in environmental cleanup efforts. In studying the mechanism of one such enzyme, scientists synthesized an irreversible inhibitor with the specific structure shown below (the enantiomer was not an inhibitor).
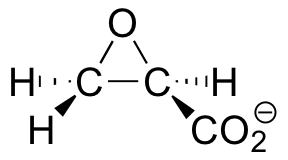
Mass spectroscopy revealed that the enzyme, after treatment with this compound, is alkylated at the carboxy-terminal proline residue.
a) Draw two possible enzyme inactivation mechanisms showing two possible proline alkylation products. Explain why your two mechanisms lead to two different products.
b) What do the findings of this study suggest about the mechanism of dehalogenation? Just state your answer in words.
C9.5: Glucosidase enzymes catalyze the cleavage of glucose-glucose bonds (R in the figure below is a second glucose molecule). Many glucosidases have two critical Asp residues in the active site, positioned above and below the site of hydrolysis:
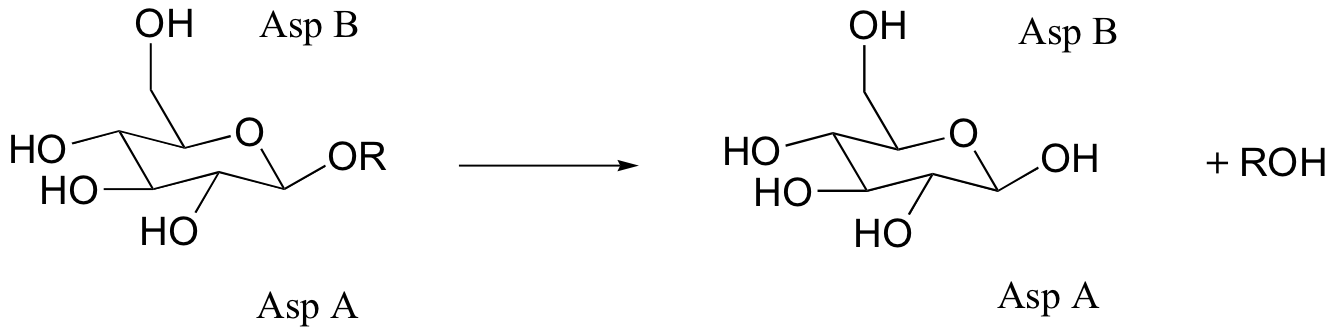
When Asp A was mutated to alanine, no enzymatic activity was observed when the enzyme was incubated with the substrate. When azide ion was added to the buffer, however, an azido-glucose product was observed (azide, N3-, is an excellent nucleophile).
a) Propose a mechanism for the normal hydrolysis reaction (with wild-type enzyme), including the role played by the two asparates.
b) Explain the formation of, and predict the structure of the azido-glucose product.
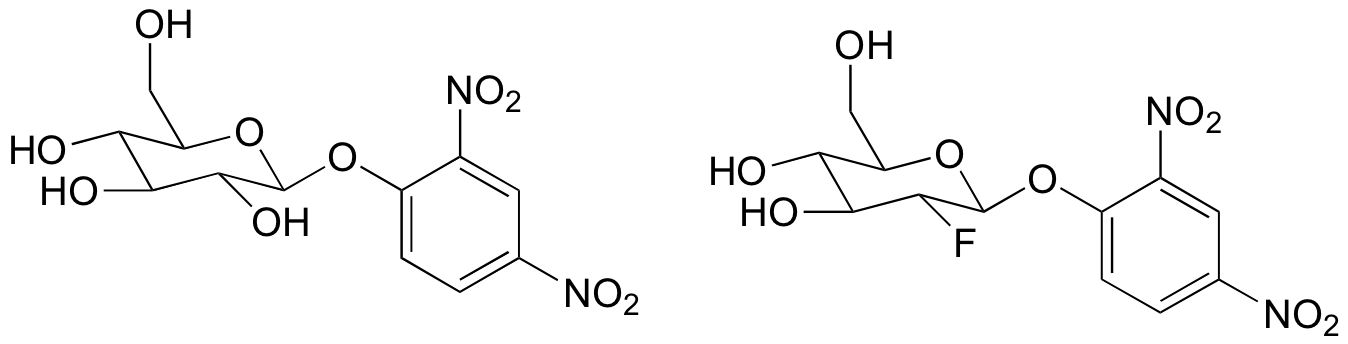
c) The substrate analogs shown below were tested with glucosidase. Why do you think they chose the dinitrophenol group in these molecules? What do you think researchers might have been trying to learn with this experiment?