5.9: Problems for Chapter 5
( \newcommand{\kernel}{\mathrm{null}\,}\)
Link to Solution Manual
P5.1: For each molecule, predict the number of signals in the 1H-NMR and the 13C-NMR spectra (do not count split peaks - eg. a quartet counts as only one signal). Assume that diastereotopic groups are non-equivalent.
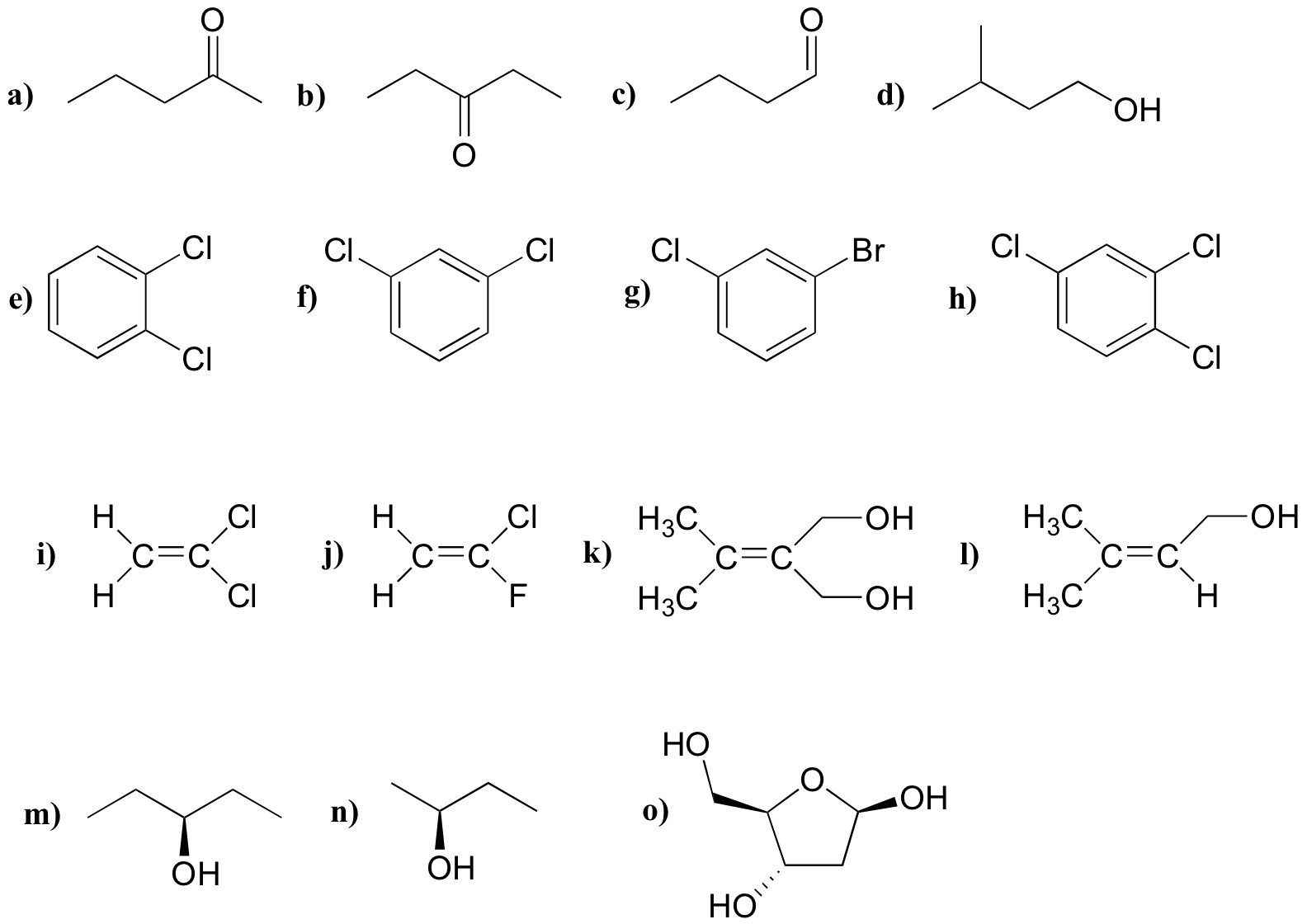
P5.2: For each of the 20 common amino acids, predict the number of signals in the proton-decoupled 13C-NMR spectrum.
P5.3: Calculate the chemical shift value (expressed in Hz, to one decimal place) of each sub-peak on the 1H-NMR doublet signal below. Do this for:
a) a spectrum obtained on a 300 MHz instrument
b) a spectrum obtained on a 100 MHz instrument
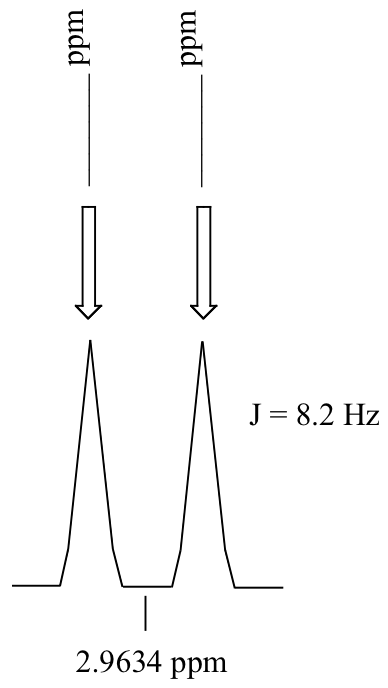
P5.4: Consider a quartet signal in an 1H-NMR spectrum obtained on a 300 MHz instrument. The chemical shift is recorded as 1.7562 ppm, and the coupling constant is J = 7.6 Hz. What is the chemical shift, expressed to the nearest 0.1 Hz, of the furthest downfield sub-peak in the quartet? What is the resonance frequency (again expressed in Hz) of this sub-peak?)
P5.5: One easily recognizable splitting pattern for the aromatic proton signals from disubstituted benzene structures is a pair of doublets. Does this pattern indicate ortho, meta, or para substitution?
P5.6 :Match spectra below to their corresponding structures A-F.
Structures:
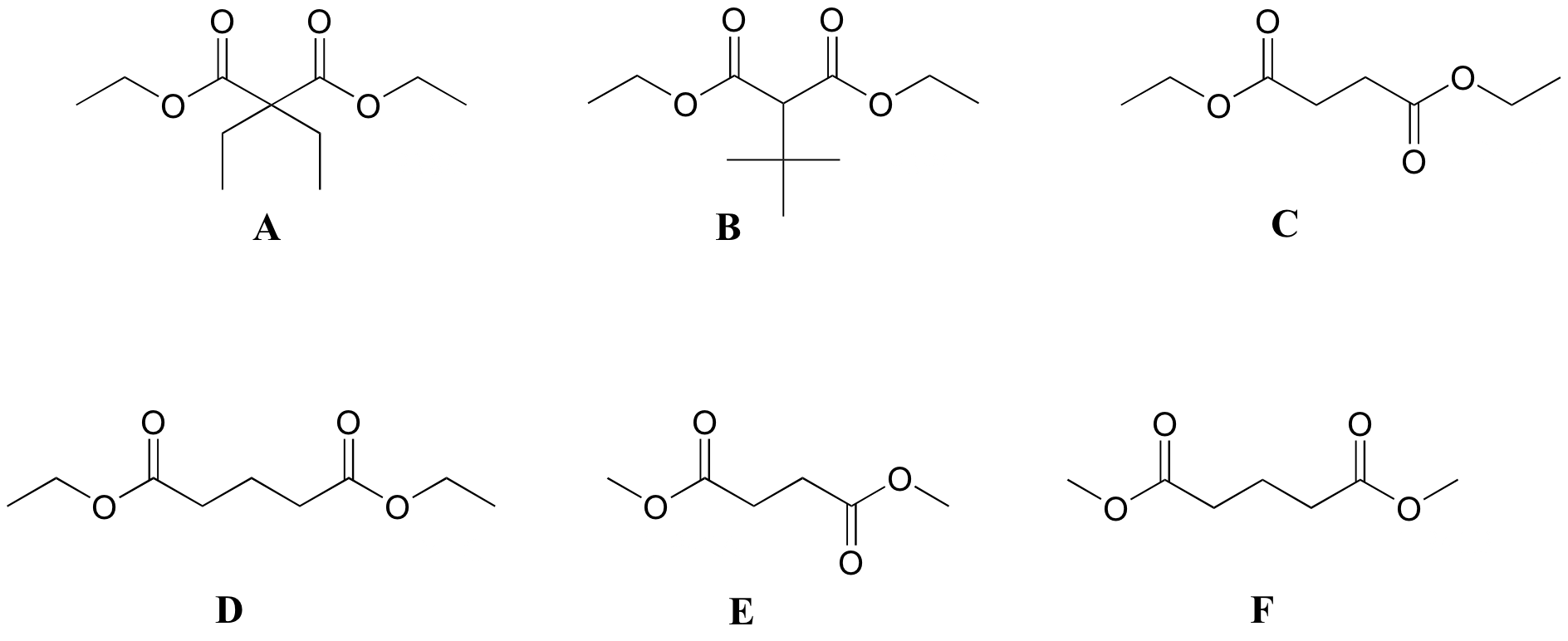
Spectrum 1
| δ | splitting | integration |
| 4.13 | q | 2 |
| 2.45 | t | 2 |
| 1.94 | quintet | 1 |
| 1.27 | t | 3 |
Spectrum 2
| δ | splitting | integration |
| 3.68 | s | 3 |
| 2.99 | t | 2 |
| 1.95 | quintet | 1 |
Spectrum 3
| δ | splitting | integration |
| 4.14 | q | 1 |
| 2.62 | s | 1 |
| 1.26 | t | 1.5 |
Spectrum 4
| δ | splitting | integration |
| 4.14 | q | 4 |
| 3.22 | s | 1 |
| 1.27 | t | 6 |
| 1.13 | s | 9 |
Spectrum 5
| δ | splitting | integration |
| 4.18 | q | 1 |
| 1.92 | q | 1 |
| 1.23 | t | 1.5 |
| 0.81 | t | 1.5 |
Spectrum 6
| δ | splitting | integration |
| 3.69 | s | 1.5 |
| 2.63 | s | 1 |
P5.7: Match spectra 7-12 below to their corresponding structures G-L .
Structures:

Spectrum 7:
| δ | splitting | integration |
| 9.96 | d | 1 |
| 5.88 | d | 1 |
| 2.17 | s | 3 |
| 1.98 | s | 3 |
Spectrum 8:
| δ | splitting | integration |
| 9.36 | s | 1 |
| 6.55 | q | 1 |
| 2.26 | q | 2 |
| 1.99 | d | 3 |
| 0.96 | t | 3 |
Spectrum 9:
| δ | splitting | integration |
| 9.57 | s | 1 |
| 6.30 | s | 1 |
| 6.00 | s | 1 |
| 1.84 | s | 3 |
Spectrum 10:
| δ | splitting | integration |
| 9.83 | t | 1 |
| 2.27 | d | 2 |
| 1.07 | s | 9 |
Spectrum 11:
| δ | splitting | integration |
| 9.75 | t | 1 |
| 2.30 | dd | 2 |
| 2.21 | m | 1 |
| 0.98 | d | 6 |
Spectrum 12:
| δ | splitting | integration |
| 8.08 | s | 1 |
| 4.13 | t | 2 |
| 1.70 | m | 2 |
| 0.96 | t | 3 |
P5.8: Match the 1H-NMR spectra 13-18 below to their corresponding structures M-R .
Structures:
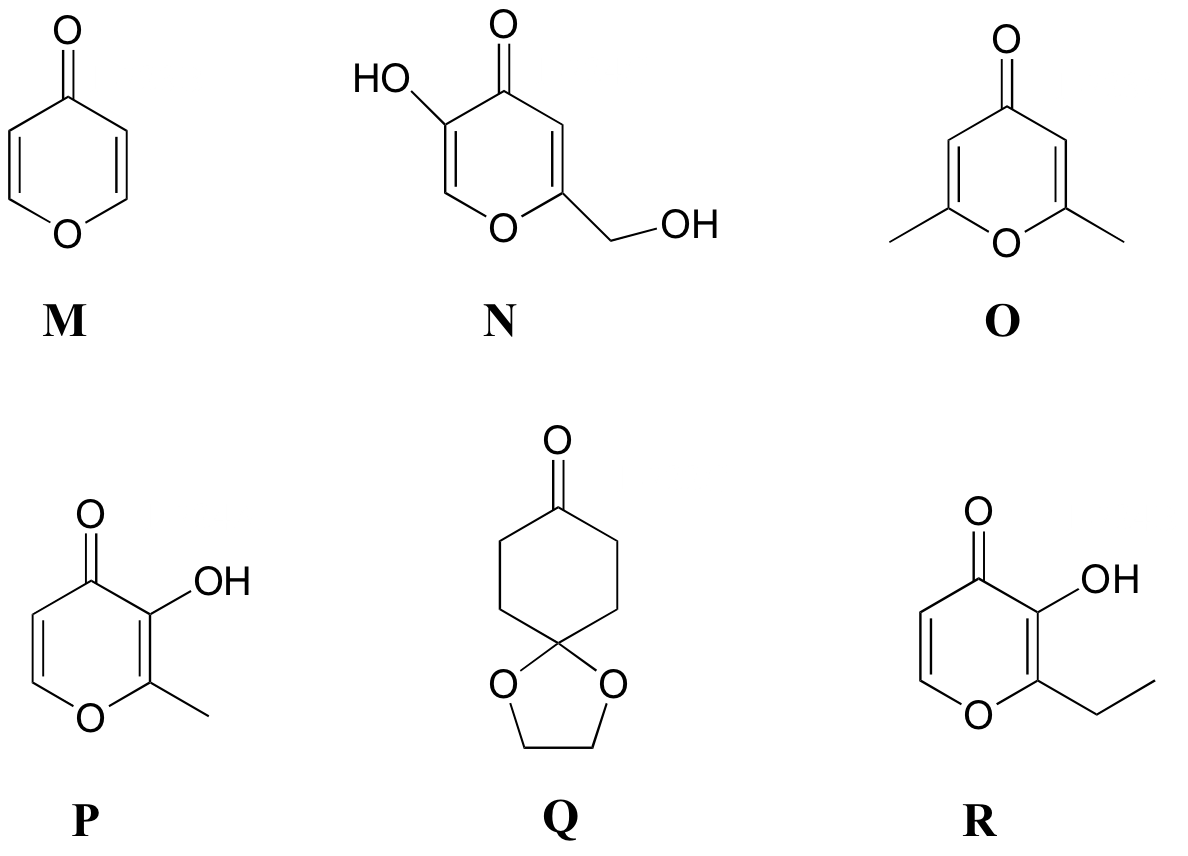
Spectrum 13:
| δ | splitting | integration |
| 8.15 | d | 1 |
| 6.33 | d | 1 |
Spectrum 14: 1-723C (structure O)
| δ | splitting | integration |
| 6.05 | s | 1 |
| 2.24 | s | 3 |
Spectrum 15:
| δ | splitting | integration |
| 8.57 | s (b) | 1 |
| 7.89 | d | 1 |
| 6.30 | d | 1 |
| 2.28 | s | 3 |
Spectrum 16:
| δ | splitting | integration |
| 9.05 | s (b) | 1 |
| 8.03 | s | 1 |
| 6.34 | s | 1 |
| 5.68 | s (b) | 1 |
| 4.31 | s | 2 |
Spectrum 17:
| δ | splitting | integration |
| 7.76 | d | 1 |
| 7.57 | s (b) | 1 |
| 6.44 | d | 1 |
| 2.78 | q | 2 |
| 1.25 | t | 3 |
Spectrum 18:
| δ | splitting | integration |
| 4.03 | s | 1 |
| 2.51 | t | 1 |
| 2.02 | t | 1 |
P5.9: Match the 1H-NMR spectra 19-24 below to their corresponding structures S-X.
Structures:

Spectrum 19:
| δ | splitting | integration |
| 9.94 | s | 1 |
| 7.77 | d | 2 |
| 7.31 | d | 2 |
| 2.43 | s | 3 |
Spectrum 20:
| δ | splitting | integration |
| 10.14 | s | 2 |
| 8.38 | s | 1 |
| 8.17 | d | 2 |
| 7.75 | t | 1 |
Spectrum 21:
| δ | splitting | integration |
| 9.98 | s | 1 |
| 7.81 | d | 2 |
| 7.50 | d | 2 |
Spectrum 22:
| δ | splitting | integration |
| 7.15-7.29 | m | 2.5 |
| 2.86 | t | 1 |
| 2.73 | t | 1 |
| 2.12 | s | 1.5 |
Spectrum 23:
| δ | splitting | integration |
| 7.10 | d | 1 |
| 6.86 | d | 1 |
| 3.78 | s | 1.5 |
| 3.61 | s | 1 |
| 2.12 | s | 1.5 |
Spectrum 24:
| δ | splitting | integration |
| 7.23-7.30 | m | 1 |
| 3.53 | s | 1 |
P5.10: Match the 1H-NMR spectra 25-30 below to their corresponding structures AA-FF.
Structures:
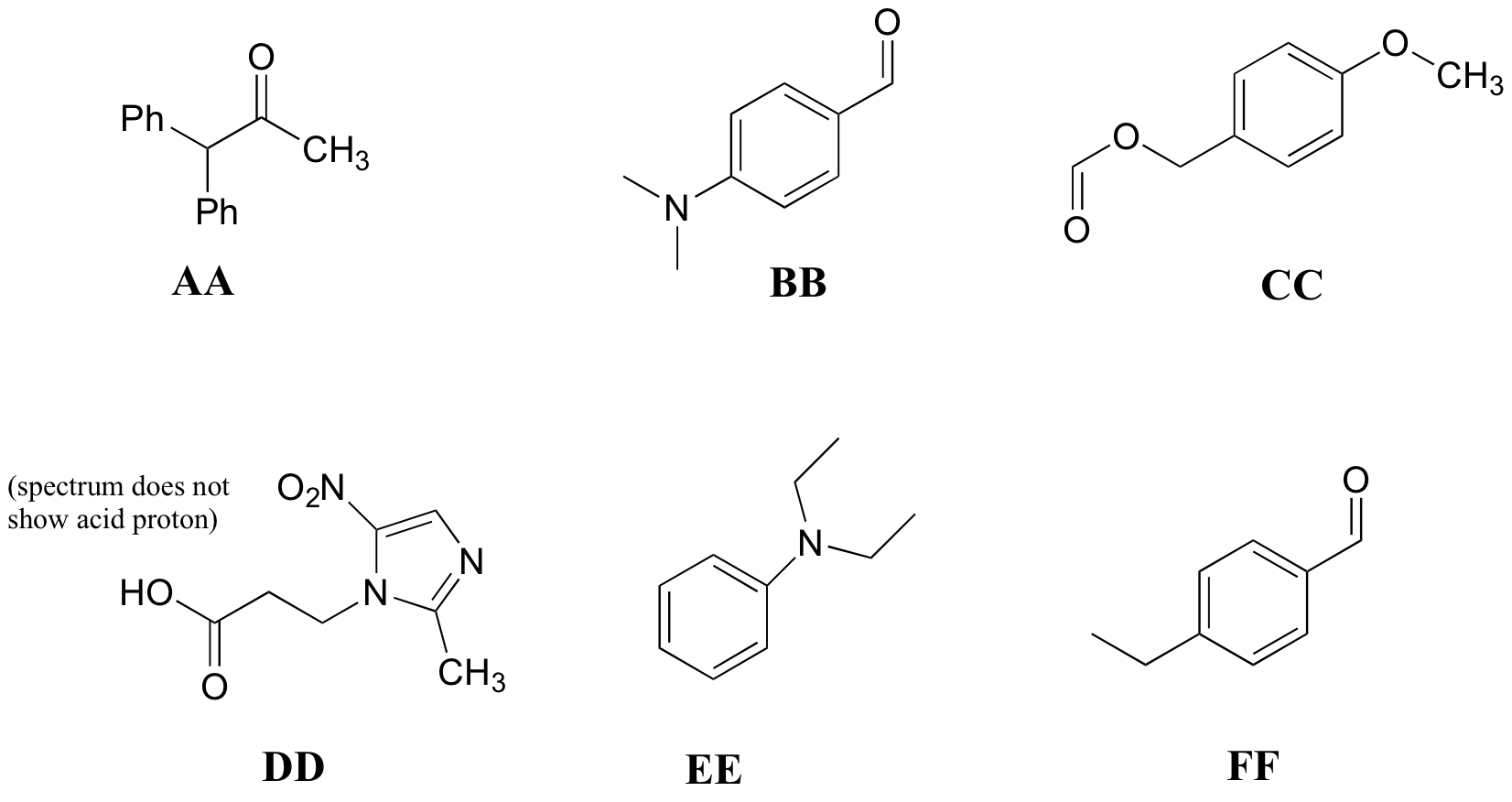
Spectrum 25:
| δ | splitting | integration |
| 9.96 | s | 1 |
| 7.79 | d | 2 |
| 7.33 | d | 2 |
| 2.72 | q | 2 |
| 1.24 | t | 3 |
Spectrum 26:
| δ | splitting | integration |
| 9.73 | s | 1 |
| 7.71 | d | 2 |
| 6.68 | d | 2 |
| 3.06 | s | 6 |
Spectrum 27:
| δ | splitting | integration |
| 7.20-7.35 | m | 10 |
| 5.12 | s | 1 |
| 2.22 | s | 3 |
Spectrum 28:
| δ | splitting | integration |
| 8.08 | s | 1 |
| 7.29 | d | 2 |
| 6.87 | d | 2 |
| 5.11 | s | 2 |
| 3.78 | s | 3 |
Spectrum 29:
| δ | splitting | integration |
| 7.18 | d | 1 |
| 6.65 | m | 1.5 |
| 3.2 | q | 2 |
| 1.13 | t | 3 |
Spectrum 30:
| δ | splitting | integration |
| 8.32 | s | 1 |
| 4.19 | t | 2 |
| 2.83 | t | 2 |
| 2.40 | s | 3 |
P5.11: Match the 1H-NMR spectra 31-36 below to their corresponding structures GG-LL
Structures:
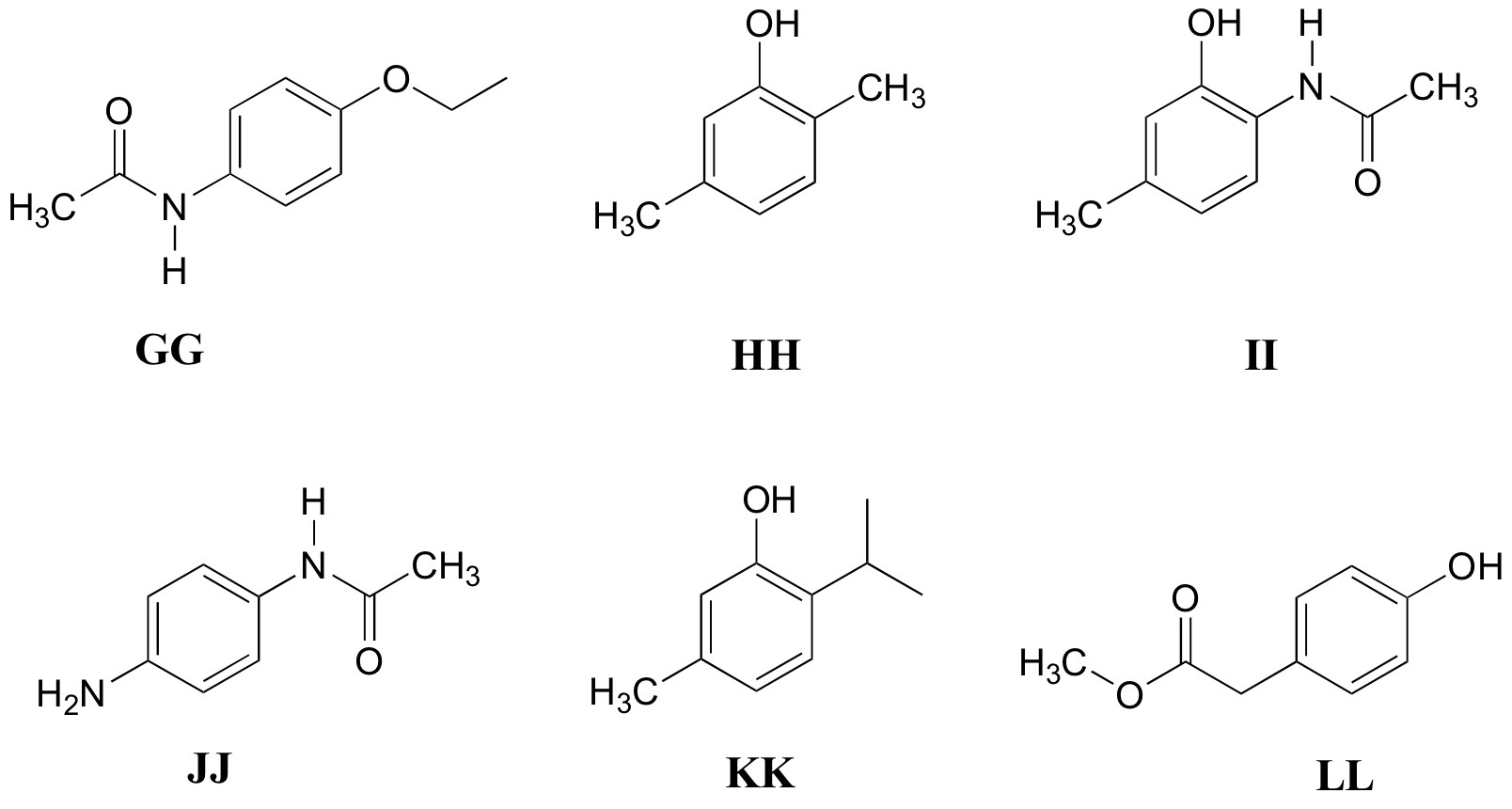
Spectrum 31:
| δ | splitting | integration |
| 6.98 | d | 1 |
| 6.64 | d | 1 |
| 6.54 | s | 1 |
| 4.95 | s | 1 |
| 2.23 | s | 3 |
| 2.17 | s | 3 |
Spectrum 32:
| δ | splitting | integration |
| 7.08 | d | 1 |
| 6.72 | d | 1 |
| 6.53 | s | 1 |
| 4.81 | s | 1 |
| 3.15 | 7-tet | 1 |
| 2.24 | s | 3 |
| 1.22 | d | 6 |
Spectrum 33:
| δ | splitting | integration |
| 7.08 | d | 2 |
| 6.71 | d | 2 |
| 6.54 | s | 1 |
| 3.69 | s | 3 |
| 3.54 | s | 2 |
Spectrum 34:
| δ | splitting | integration |
| 9.63 | s | 1 |
| 7.45 | d | 2 |
| 6.77 | d | 2 |
| 3.95 | q | 2 |
| 2.05 | s | 3 |
| 1.33 | t | 3 |
Spectrum 35:
| δ | splitting | integration |
| 9.49 | s | 1 |
| 7.20 | d | 2 |
| 6.49 | d | 2 |
| 4.82 | s | 2 |
| 1.963 | s | 3 |
Spectrum 36:
| δ | splitting | integration |
| 9.58 | s(b) | 1 |
| 9.31 | s | 1 |
| 7.36 | d | 1 |
| 6.67 | s | 1 |
| 6.55 | d | 1 |
| 2.21 | s | 3 |
| 2.11 | s | 3 |
P5.12: Use the NMR data given to deduce structures.
a ) Molecular formula: C5H8O
1H-NMR:
| δ | splitting | integration |
| 9.56 | s | 1 |
| 6.25 | d (J~1 Hz) | 1 |
| 5.99 | d (J~1 Hz) | 1 |
| 2.27 | q | 2 |
| 1.18 | t | 3 |
13C-NMR
| δ | DEPT |
| 194.60 | CH |
| 151.77 | C |
| 132.99 | CH2 |
| 20.91 | CH2 |
| 11.92 | CH3 |
b) Molecular formula: C7H14O2
1H-NMR:
| δ | splitting | integration |
| 3.85 | d | 2 |
| 2.32 | q | 2 |
| 1.93 | m | 1 |
| 1.14 | t | 3 |
| 0.94 | d | 6 |
13C-NMR
| δ | DEPT |
| 174.47 | C |
| 70.41 | CH2 |
| 27.77 | CH |
| 27.64 | CH2 |
| 19.09 | CH3 |
| 9.21 | CH3 |
c) Molecular formula: C5H12O
1H-NMR:
| δ | splitting | integration |
| 3.38 | s | 2H |
| 2.17 | s | 1H |
| 0.91 | s | 9H |
13C-NMR
| δ | DEPT |
| 73.35 | CH2 |
| 32.61 | C |
| 26.04 | CH3 |
d) Molecular formula: C10H12O
1H-NMR:
| δ | splitting | integration |
| 7.18-7.35 | m | 2.5 |
| 3.66 | s | 1 |
| 2.44 | q | 1 |
| 1.01 | t | 1.5 |
13C-NMR
| δ | DEPT |
| 208.79 | C |
| 134.43 | C |
| 129.31 | CH |
| 128.61 | CH |
| 126.86 | CH |
| 49.77 | CH2 |
| 35.16 | CH2 |
| 7.75 | CH3 |
P5.13:
13C-NMR data is given for the molecules shown below. Complete the peak assignment column of each NMR data table.
a)
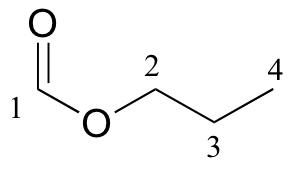
| δ | DEPT | carbon # |
| 161.12 | CH |
|
| 65.54 | CH2 |
|
| 21.98 | CH2 |
|
| 10.31 | CH3 |
|
b)
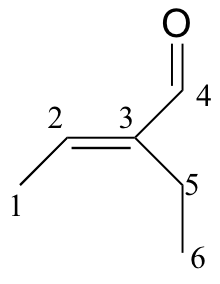
| δ | DEPT | carbon # |
| 194.72 | C |
|
| 149.10 | C |
|
| 146.33 | CH |
|
| 16.93 | CH2 |
|
| 14.47 | CH3 |
|
| 12.93 | CH3 |
|
c)
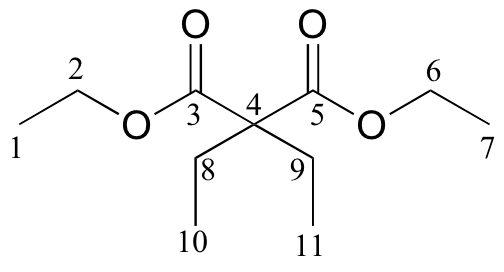
| δ | DEPT | carbon # |
| 171.76 | C |
|
| 60.87 | CH2 |
|
| 58.36 | C |
|
| 24.66 | CH2 |
|
| 14.14 | CH3 |
|
| 8.35 | CH3 |
|
d)
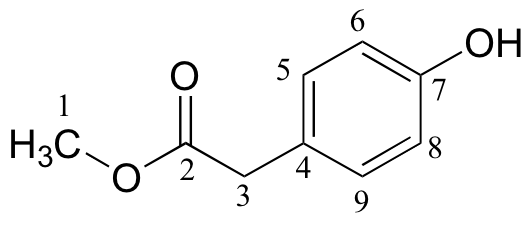
| δ | DEPT | carbon # |
| 173.45 | C |
|
| 155.01 | C |
|
| 130.34 | CH |
|
| 125.34 | C |
|
| 115.56 | CH |
|
| 52.27 | CH3 |
|
| 40.27 | CH2 |
|
e)
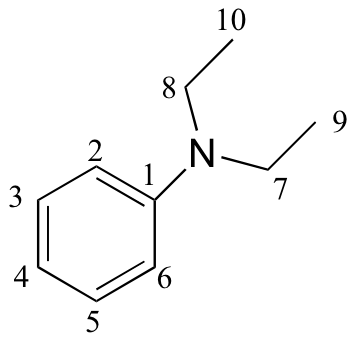
| δ | DEPT | carbon # |
| 147.79 | C |
|
| 129.18 | CH |
|
| 115.36 | CH |
|
| 111.89 | CH |
|
| 44.29 | CH2 |
|
| 12.57 | CH3 |
|
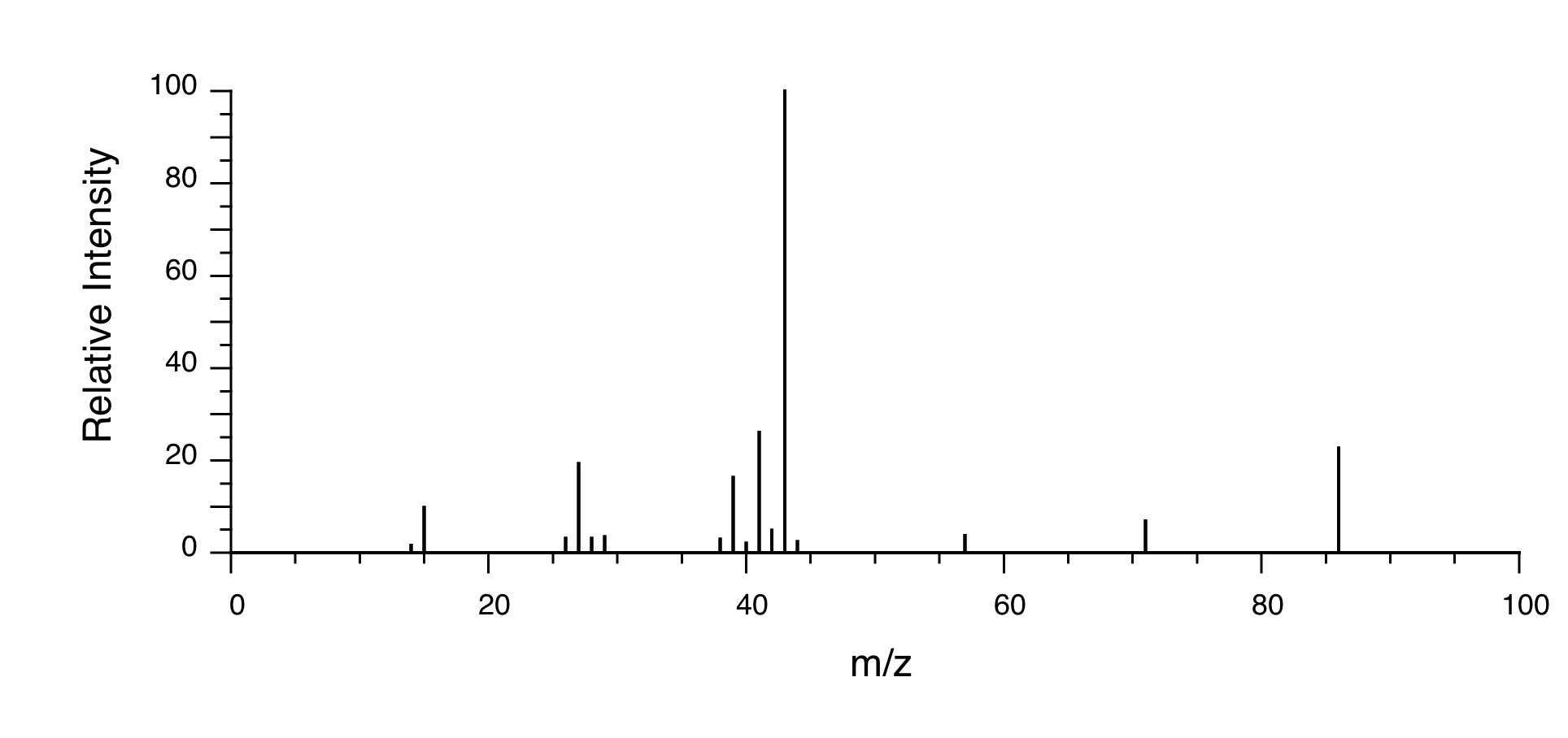
P5.14: You obtain the following data for an unknown sample. Deduce its structure.
1H-NMR:
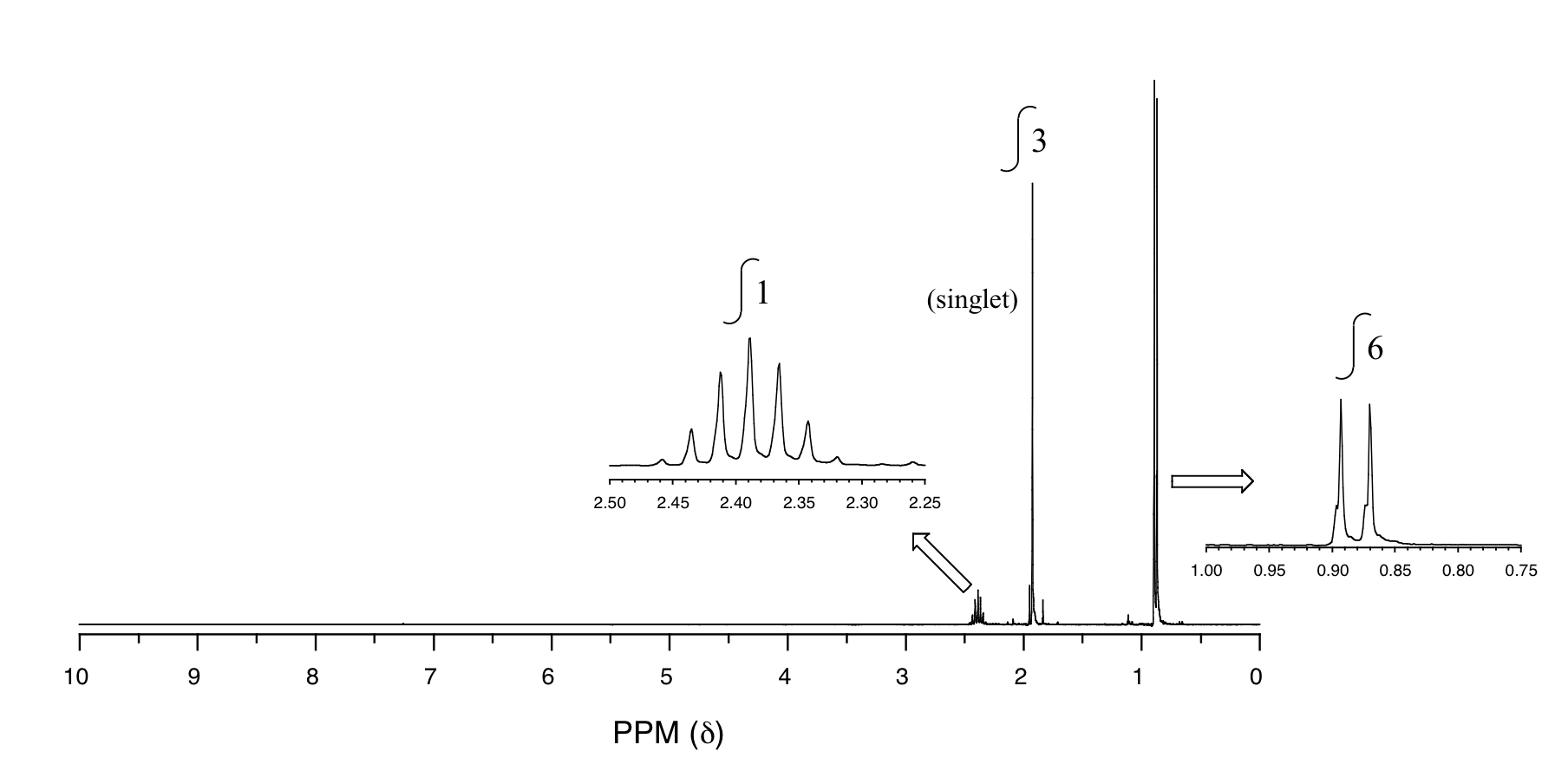
13C-NMR:
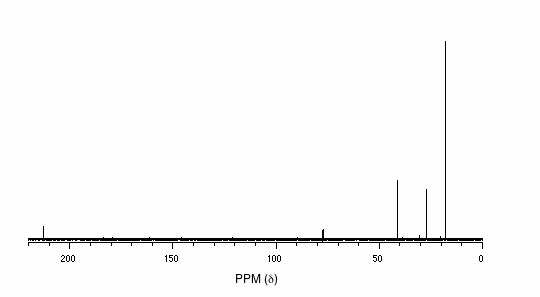
Mass Spectrometry:
P5.15:You take a 1H-NMR spectrum of a sample that comes from a bottle of 1-bromopropane. However, you suspect that the bottle might be contaminated with 2-bromopropane. The NMR spectrum shows the following peaks:
| δ | splitting | integration |
| 4.3 | septet | 0.0735 |
| 3.4 | triplet | 0.661 |
| 1.9 | sextet | 0.665 |
| 1.7 | doublet | 0.441 |
| 1.0 | triplet | 1.00 |
How badly is the bottle contaminated? Specifically, what percent of the molecules in the bottle are 2-bromopropane?
Challenge problems
C5.1: All of the 13C-NMR spectra shown in this chapter include a signal due to CDCl3, the solvent used in each case. Explain the splitting pattern for this signal.
C5.2: Researchers wanted to investigate a reaction which can be catalyzed by the enzyme alcohol dehydrogenase in yeast. They treated 4'-acylpyridine (1) with living yeast, and isolated the alcohol product(s) (some combination of 2A and 2B).
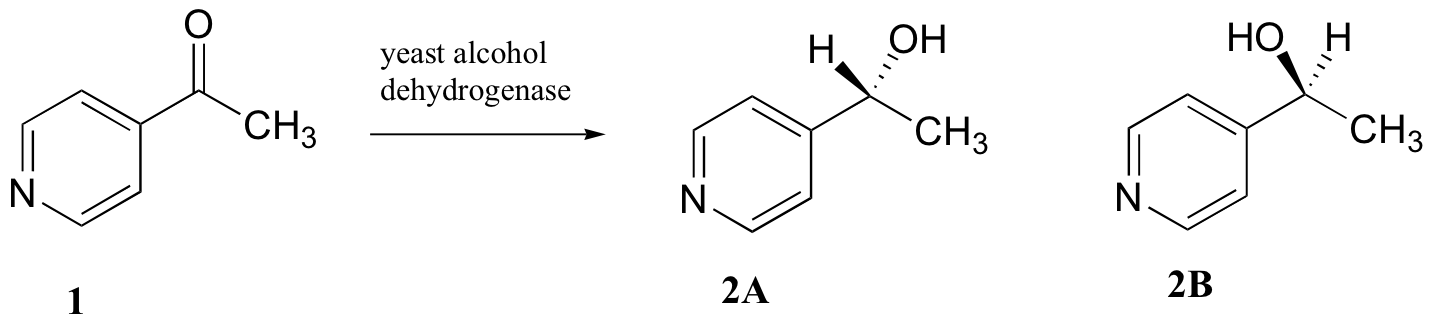
a) Will the products 2A and 2B have identical or different 1H-NMR spectra? Explain.
b) Suggest a 1H-NMR experiment that could be used to determine what percent of starting material (1) got turned into product (2A and 2B).
c) With purified 2A/2B, the researchers carried out the subsequent reaction shown below to make 3A and 3B, known as 'Mosher's esters'. Do 3A and 3B have identical or different 1H-NMR spectra? Explain.
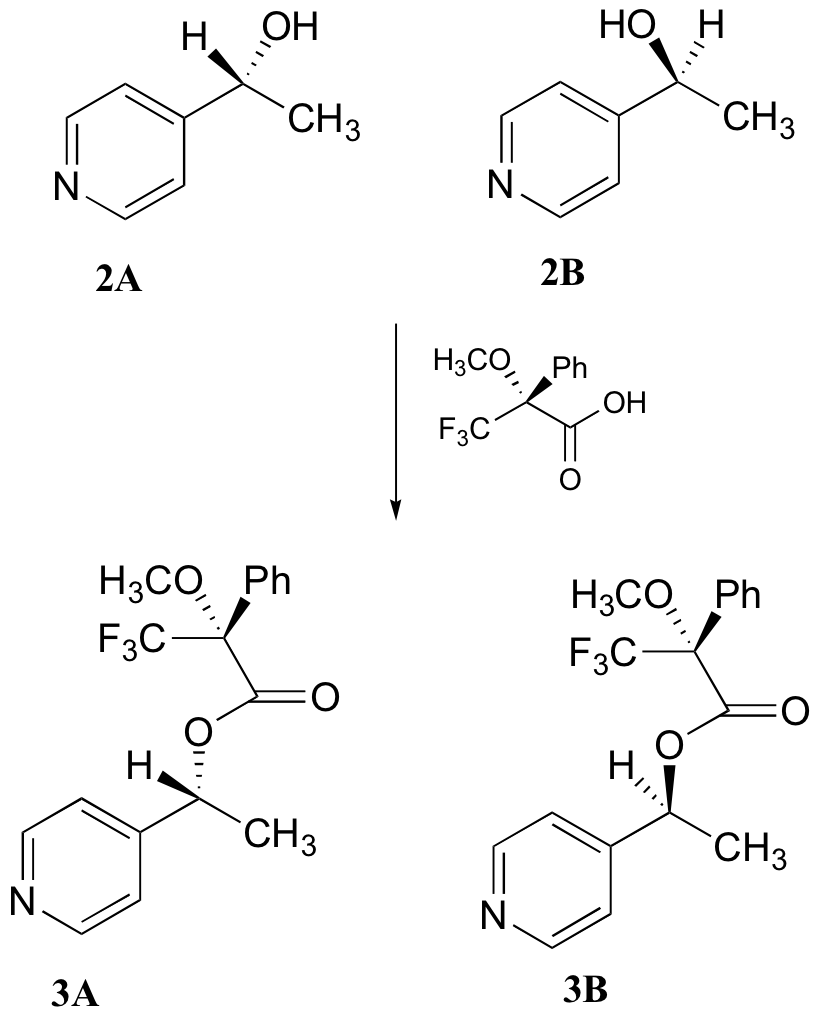
d) Explain, very specifically, how the researchers could use 1H-NMR to determine the relative amounts of 2A and 2B formed in the reaction catalyzed by yeast enzyme.


